Decisive bioprocess parameters such as biomass and product concentration or metabolite concentrations in the medium are often not directly measurable, requiring offline sampling which does not provide immediate results. Despite the development of new measuring systems and probes, not every parameter can always be measured directly online. This can be remedied by soft-sensors (sometimes also called software sensors), which calculate the desired parameter based on a model or a simple correlation of known measured values.
What are soft-sensors good for and what are their advantages? The crucial point is that soft-sensors do not provide “new” data but help the user to put the existing data into a more useful and understandable format by combining different measurable data into one parameter.
This new “soft-sensor measured value” can then be used for process control and automation. A soft-sensor is as flexible as the underlying model and can be adapted to the process, reactor and organism used. Another advantage is that existing infrastructure can be utilized with a corresponding bioprocess software, eiminating the need for new hardware purchases.
How can you use soft-sensors?
Which parameters can be determined using soft-sensors and how complex will the procedure be? In order to answer this question, I would like to divide the soft-sensors into four categories.
1. Simple mathematical conversions help you to evaluate or compare bioprocess data
For example the impeller tip speed can be calculated from the stirrer speed using a simple conversion, which is commonly used to compare bioreactors with different stirrers. If the media viscosity is known, the Reynolds number can also be determined, allowing for a statement relating to the turbulence in the bioreactor. Another typical conversion example is to calculate the carbon load imparted to the system, by multiplying the federate by the carbon content in the feed solution.
2. You can combine different measurements to obtain more information on the bioprocess
A typical example is the oxygen uptake rate (OUR). The combination of dissolved oxygen concentration and input gas flows allows the researcher to measure OUR, which can then be used to estimate the biomass growth. The same can likewise be achieved by means of exhaust gas analysis, which can be used to calculate the carbon dioxide evolution rate (CER). Both can subsequently be combined to obtain the respiratory quotient (RQ), which shows that soft-sensors can also be input variables for other soft-sensors. Another example would be to calculate the cultivation volume, by integrating the pump signals over time, instead of measuring it by a scale.
3. You can detect correlations with offline determined values
Certain values are measured offline. Some of these offline values can also be calculated from suitable online measurements if a correlation formula is known. Soft-sensors can be used to determine the correlation factors, and to provide a real time calculation for the offline values equally. A typical example is the biomass concentration, which is typically determined from offline samples. However, it can be correlated with the signal of a conductivity or permittivity probe or the measured backscattered light in order to obtain an online estimation.
4. You can integrate different bioprocess engineering models
When charaterizing a bioreactor in process engineering terms, first the kLa value, mixing time or power output at different stirrer speeds, filling volumes and gassing rates will need to be measured. With this data a model can be formed and be integrated into the process as a soft-sensor. A similar approach would be to integrate of an offline kinetic model (e.g. a simple Monod model), which could then be correlated with an online measurable signal and integrated into the process control.
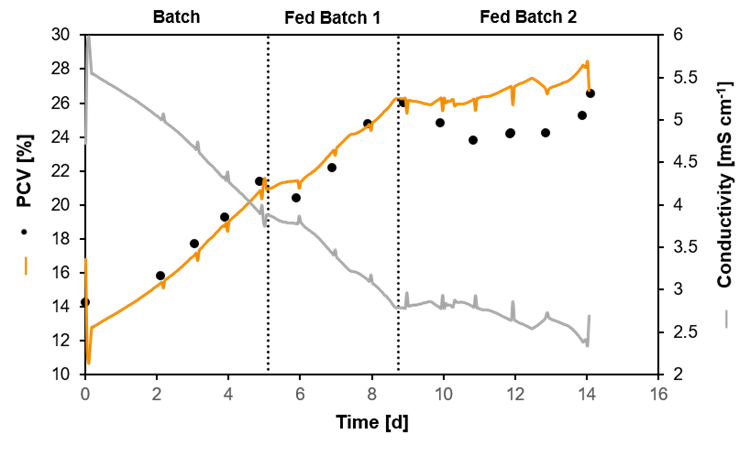
Packed Cell Volume (PCV) estimated by a soft-sensor. The measured conductivity was used to predict the PCV in a heterotrophic cultivation of plant cells. The black dots show the PCV evaluated by offline samples.
How can you create soft-sensors?
As described, the possibilities for soft-sensors are manifold. But what information is needed to create a soft-sensor? The most important is certainly the mathematical model. For many applications, this model is a simple, e.g. linear, correlation between inputs and outputs. You rarely have to be really creative; many models have already been predefined and verified sufficiently in the scientific literature. In addition, there are often several variants published for the same measurand. This makes it easier to fit a soft-sensor to the inputs available at your bioreactor.
For the creation of a new soft-sensor, the inputs, outputs and coefficients must first be defined. The latter can also be adjusted during the running process, for example if the volume in a fed batch changes or a temperature shift in the production phase is detected.
The actual soft-sensor is then scripted. In most platforms, this is done a programming language such as the widely used C#. This part frightens many users, but it is not as complicated as it looks. Most bioprocessing software come with instructions and examples on how to create soft-sensors. Once created, soft-sensors can also be used as templates for others. In addition, the programming environment tests the syntax and points out errors before the soft-sensor is even used. In order to avoid errors, it is recommended to use the soft-sensor with historical data and to check the results before applying it to new processes.

Generic procedure to develop a soft-sensor
Summary
In summary it can be said that soft-sensors offer excellent possibilities to increase the process understanding and to facilitate and automate the evaluation. It is important to choose a suitable, preferably already verified model which fits to the already existing hardware sensors. The actual scripting may seem challenging at first sight, but with examples, instruction and support, it is easy to increase your system’s overall capability.
I think it is worth trying. Even with a bare minimum of programming, one can unlock the full potential of a cultivation system – without having to buy new and expensive hardware. At least for me, this is not only sensible, it is also good fun.
Rüdiger Maschke is graduated as Dipl.-Ing. in chemical engineering at the TU Dresden and now working at the Zurich University of Applied Sciences (ZHAW) within the competence centre of biochemical engineering and cell culture techniques. He devotes his research to reaction kinetics and simulations specially for cell cultures as well as bio process engineering and process transfer (scale up/down).

Amit Damor
Lab technician software